Bioinformatics is the subject of today's science. Various Bioinformatics tools are available online. Scholars like to search mainly NCBI blastp, NCBI blastn, NCBI ftp site, NCBI nucleotide blast, primer blast NCBI, blast 2, blast search, and various other keywords related to Bioinformatics tools.
In this post, you will learn:
- How to download/copy genome sequence
- How to change genome format (FASTA Format)
- How to run BLAST on gene/protein sequence
Download NCBI gene sequence
Search NCBI on the google page.
Click on National Center for Biotechnology
Information
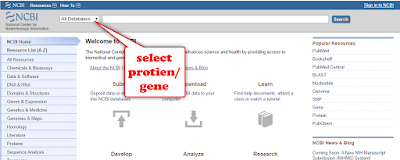
Select gene/protein
from “all database" option
Write the name of the protein/gene on the box and click on "Search" as shown above.
All reports, related to the topic will appear, click on
desire report to get a complete version.
Copy the sequence given on the last
of the page in the heading of ORIGIN by pressing Ctrl+C.
Note: Don’t
copy "digits" and "//" as shown below.
Save this sequence by pasting in a word file. Use this sequence for the required purposes.
See Also: Best Batteries 👈
How to get FASTA format
To get FASTA format, follow the procedure discussed above and click on “FASTA” on the top of page.
 |
| Sequence after applying FASTA |
How to run BLAST
Search NCBI on the google page.
Click on National Center for Biotechnology Information
Click on Resource List (A-Z)
Click on Basic Local
Alignment Search Tool (BLAST)
Paste the sequence in the following box and click on "Blast"
Color
map of protein/gene sequence (Graphic summary) will appear with a complete
description. As shown below: `
Redline shows that only with 2
organisms it has 100% similarity.
The purple line shows that with a few organisms
it has less than 80% similarity.
Green shows that with some organisms it
has similarity up to 50%
Blue shows that with most organism it
has a similarity between 40 to 50%
Black shows that with some organism it
shows a similarity less than 40%.










Post A Comment:
0 comments so far,add yours